The accreditors of this session require that you periodically check in to verify that you are still attentive. Please click the button below to indicate that you are.
DEVELOPMENT OF EXPLAINABLE COMPUTER-AIDED DIAGNOSIS USING REPRODUCIBLE ANALYSIS AND INTERPRETABLE FEATURES IN COLONOSCOPY
Method: The development of CADx with XAI comprises four steps: (1) Data preparation involving radiomics, color, deep features (Dataset A: 5492 ADs and 1576 HPs) and endoscopists-assigned NBI International Colorectal Endoscopic (NICE) features including surface, vessel and color patterns (Dataset B: 120 ADs and 120 HPs) ; (2) Correlation-driven selection process to identify significant correlations between radiomics/color and deep features; (3) Regression analysis linking NICE grading to chosen deep features, thereby translating deep learning outcomes into NICE grading and encompassing feature selection; (4) Reinterpreting the selected deep features in terms of radiomics. The performance of CADx with XAI and three endoscopists was evaluated for another 300 polyps (Dataset C: 160 ADs and 140 HPs).
Result: An initial 2,048 deep features were identified, which was narrowed down to 103 in the second screening, and finally, 14 were selected in the final screening. Similarly, 24 out of 93 radiomics features were selected. No color features were selected during feature screening. A comparative evaluation revealed that the CADx with XAI model exhibited comparable accuracy to deep learning models while offering interpretable insights. In the test set (Dataset C), the CADx with XAI system showed 0.946 of AUC, 0.883 of ACC, 0.888 of sensitivity, 0.879 of specificity, 0.893 of positive predictive value (PPV), and 0.872 of negative predictive value (NPV) for classification of HP and AD. The system met the Simple Optical Diagnosis Accuracy (SODA) criteria and, under conditions of high confidence, also achieved the Preservation and Incorporation of Valuable Endoscopic Innovations (PIVI) NPV thresholds. A comparative study between the XAI-inferred NICE grading and endoscopists’ NICE assessments revealed a strong correlation (Spearman's 0.640, Kendall's 0.494), indicating high concordance and validating the model's clinical relevance. Furthermore, additional explanatory values using radiomics were presented.
Conclusion: This study introduced a novel CADx with XAI system that combines radiomics and deep features to enhance the transparency and understanding of the optical diagnosis. This approach bridges the gap between AI predictions and clinically meaningful assessments, offering a practical solution for enhancing diagnostic accuracy and clinical decision-making.
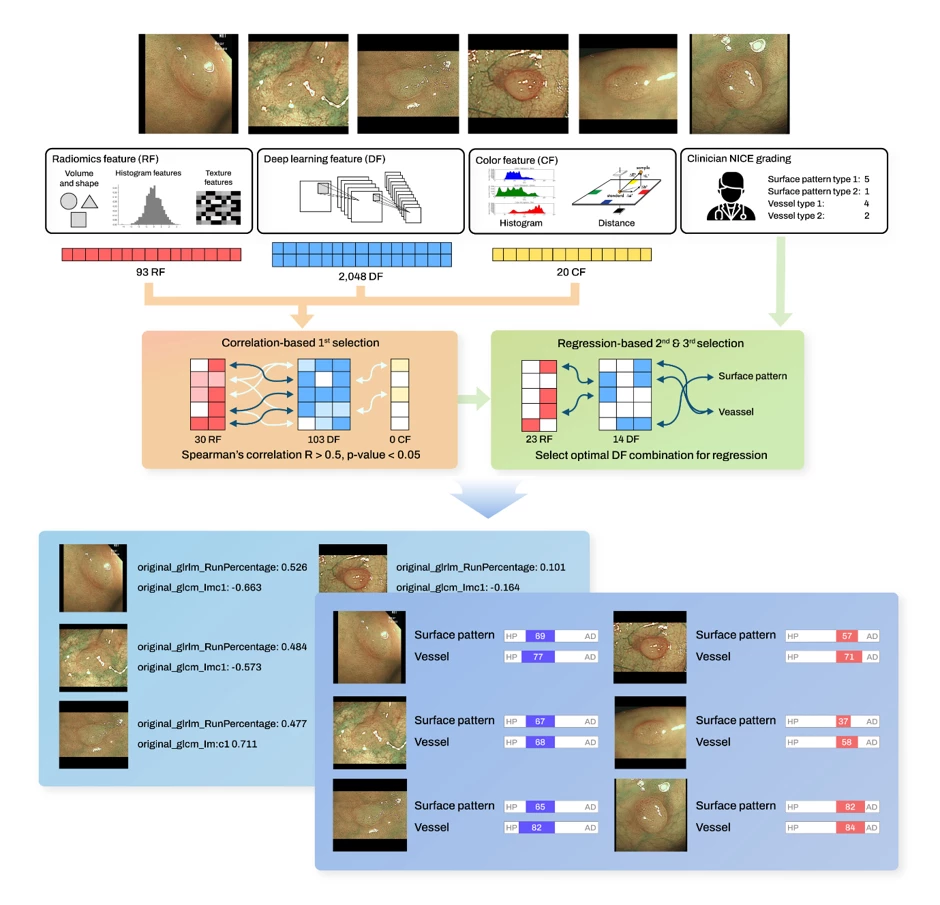
Figure 1. CADx with XAI system workflow. The CADx with XAI system comprises four integral steps: data preparation involving radiomics, color, deep features, and clinician-assigned NICE grading; a correlation-driven selection process to identify significant correlations between radiomics/color and deep features; regression analysis linking NICE grading to chosen deep features, thereby translating deep learning outcomes into NICE grading and encompassing feature selection; and finally, reinterpreting the selected deep features in terms of radiomics.
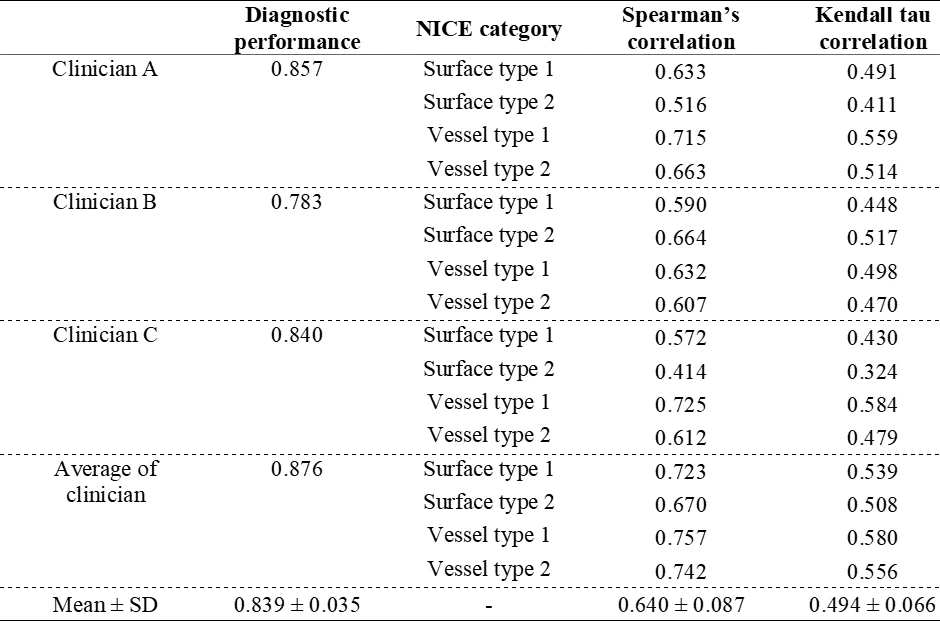
Table 1. Correlation results between the XAI NICE prediction and clinician NICE grading


