740 - OUTCOMES OF PEDIATRIC MASS SCREENING-IDENTIFIED CELIAC DISEASE
Methods: In a sub-cohort of 307 generally healthy men from a prospective cohort study established in 1986, we collected up to two pairs of stools 6 months apart, assessed MO3FA intake from two sets of 7-day diet records, and measured plasma CRP as a marker for chronic systemic inflammation at stool collection (Fig. 1A). Using metagenomic shotgun sequencing, we calculated the abundances of microbial species, functional pathways (MetaCyc), and enzyme families (ECs). We assessed the association of MO3FA intake with microbial features using MaAsLin 2 and the modifying effect by microbial species on the association of MO3FA intake with plasma CRP levels.
Results: MO3FA intake was associated with changes in specific microbial abundances (although not overall composition), with EPA and DHA showing distinct patterns from DPA (Fig. 1G). The strongest association was found between higher EPA and DHA intakes and increased abundance of Lachnospira pectinoschiza (FDR q = 0.011 for EPA and 0.007 for DHA), one of a broad set of gut microbes associated with strict anaerobicity and eubiosis(Fig. 1H). Similar associations were identified for L. pectinoschiza ECs and pathways (Fig. 1I,L). Gut microbial features modified the association between MO3FA intakes and lower plasma CRP levels, with a stronger association among participants with the presence of Eubacterium rectale (Pinteraction = 0.017 for EPA, 0.011 for DPA, 0.005 for DHA, Fig. 2B) and the absence of Phocaeicola vulgatus (Pinteraction = 0.001 for EPA and 0.008 for DHA, Fig. 2C). Both taxa are prevalent gut residents, with E. rectale again associated with strict anaerobicity and P. vulgatus known to have strain-dependent influences on immune and inflammatory responses. Similar significant interactions were found for the 4 MetaCyc pathways contributed by E. rectale (Fig. 2D,E), and 5 ECs and 1 MetaCyc pathway contributed by P. vulgatus (Fig. 2F,G). A higher level of phosphatidyl-myo-inositol alpha-mannosyltransferase (mannosyltransferase PimA) contributed by P. vulgatus, an enzyme related to the synthesis of surface-exposed lipoglycans, reversed the association of MO3FA intake and plasma CRP from a negative to positive direction (Fig. 2I).
Conclusions: Our findings support the potential role of MO3FA intake in gut microbiome composition and function, and the effect modification by the gut microbiome on the effect of MO3FA on systemic inflammation.

Figure 1. Marine omega-3 fatty acids (MO3FAs), plasma C-reactive protein (CRP), the gut microbiome, and the association between MO3FAs and the gut microbiome. Lanchnospira pectinoschiza, their enzymes and pathways are positively associated with MO3FA, eicosapentaenoic acid (EPA) and docosapentaenoic acid (DPA) but not docosahexaenoic acid (DHA). A, Study design of the study B-D, Characteristics of MO3FA intakes, and CRP levels of participants E, Principal coordinate (PCo) analyses using species-level dissimilarity. F, Taxonomic variation explained by MO3FA intakes, CRP, and covariates using permutational multivariate analysis of variance. G, Significant associations of MO3FAs with species, and the scatter plot of L. pectinoschiza (H) I,L Significant associations of MO3FAs with enzymes (I) or pathways (L) of L. pectinoschiza, and the scatter plot of EC 3.1.4.52 (J) or EC 5.2.1.8 (K).

Figure 2. The effect modification of gut microbiome on the association between Marine omega-3 fatty acids (MO3FAs) intakes and plasma C-reactive protein (CRP) levels. Eubacterium rectale, Phocaeicola vulgatus and their enzymes and pathways modify the association between MO3FA intakes and CRP level. A, The interaction of the top 10 abundant species with MO3FA intakes on the CRP level. B,C, Modification on the association between MO3FA intakes and CRP level by carriages of E. rectale (B) or P. vulgatus (C) D-G, The modification by E. rectale-contributed EC enzymes (D) and MetaCyc pathways (E), and P. vulgatus-contributed EC enzymes (F) and MetaCyc pathways (G) on the association of MO3FA intake and CRP level. H, Modification of 5 EC enzymes on the association between docosapentaenoic acid intakes and CRP level, including mannosyltransferase PimA of P. vulgatus. (I)
Methods: In recently published work, we discovered a newly evolved, cross-species compatible AAV (cc.47) that demonstrates substantially increased transduction efficiency across multiple tissues compared to the parental AAV9 capsid. Here, we tested the efficacy of intravenous and colonoscopy-guided injection of AAV.cc47 packaging a self-complementary CBh-Cre recombinase cassette to transduce the intestinal epithelium in Ai9 tdTomato fluorescent reporter mice. We created a CF mouse model carrying the W1282X nonsense mutation in Cftr using CRISPR/Cas9 gene editing and assessed Cftr loss of function in intestinal organoids using a forskolin swelling assay. Mouse embryonic fibroblasts were derived from W1282X/W1282X mice and then transduced using electroporation with an adenine base editor and sgRNA to determine editing efficiency in vitro.
Results: We found that AAV.cc47 infects intestinal stem cells with remarkably high (~ 70%) efficiency (Figure 1). We tested AAV.cc47 for targeted delivery to the colon via colonoscopy-guided injection in mice, which demonstrated moderate transduction efficiency in colonic stem cells. The W1282X Cftr mouse model recapitulated key features of human disease, including defective Cftr function based on a forskolin swelling assay in intestinal organoids. The adenine base editor successfully repaired the W1282X Cftr point mutation in vitro in mouse embryonic fibroblasts at the correct nucleotide position (Figure 2).
Conclusions: We demonstrated efficient transduction of intestinal stem cells with a newly evolved AAV that we recently published. We showed that an adenine base editor successfully repairs a CF point mutation in vitro. We will now assess the ability of an optimized AAV base editor system to repair the W1282X Cftr point mutation in our mouse model and to edit intestinal stem cells in wild-type pigs. Results from these studies will lead to the development of this gene editing technology to target intestinal disease associated with CF and other inborn genetic disorders.
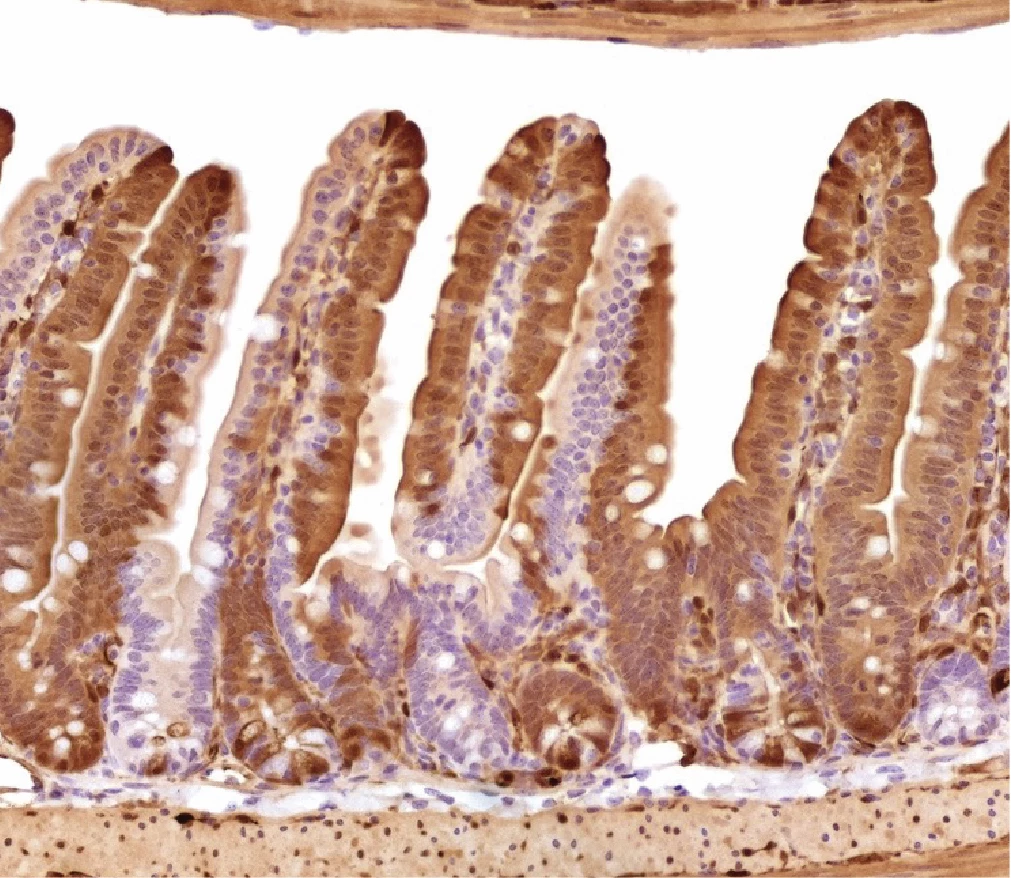
Figure 1. Intravenous delivery of AAV cc.47 results in high efficiency of intestinal epithelial transfection. Representative image of tdTomato immunohistochemistry of jejunal intestine collected one month after intravenous injection of cc.47-scCbh-Cre (1e12 vg/mouse, N=5 mice) (20X).
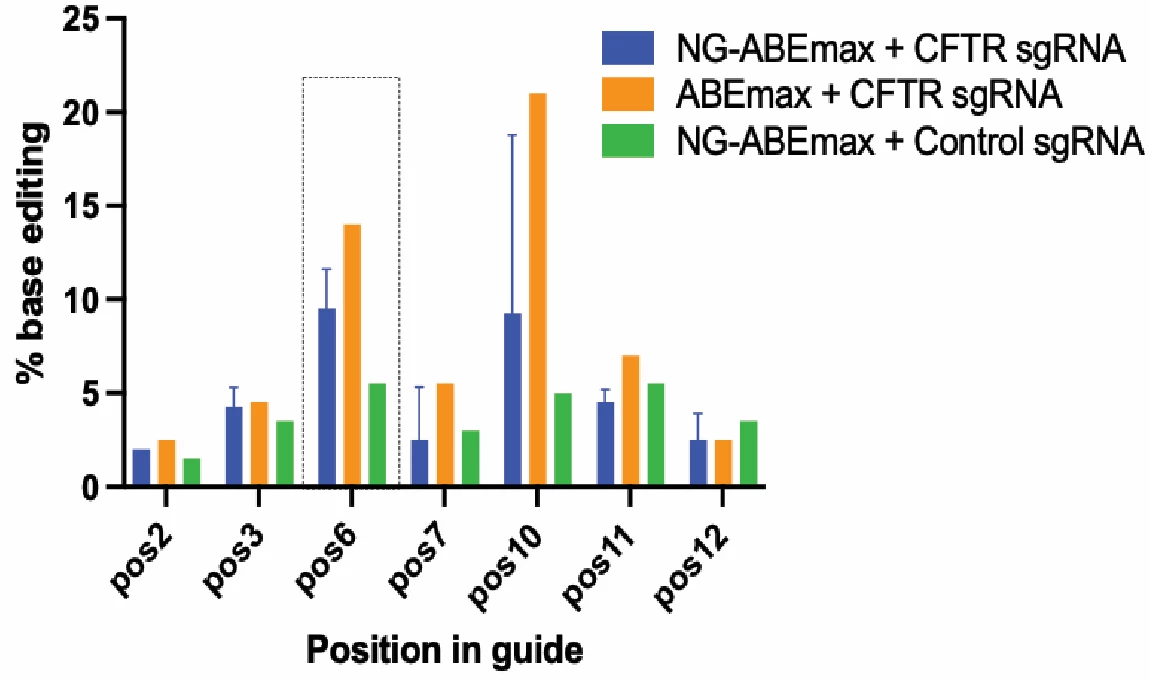
Figure 2. Successful electroporation of CftrW1282X/W1282X mouse embryonic fibroblasts with adenine base editor and sgRNA to edit the W1282X mutation. CftrW1282X/W1282X mouse embryonic fibroblasts were electroporated with NG-ABEmax plasmid with sgRNA targeting the W1282X Cftr mutation, ABEmax with sgRNA, or NG-ABEmax with a Control sgRNA. After Sanger sequencing was performed, the sequences were analyzed using EditR software. Percentage of edits was quantified at each adenosine base within the guide sequence as these are the potential sites of base editing with the adenine base editor. The mutated base is located at position 6 (denoted by the dashed box) (N=2).
The United States Preventative Service Taskforce concluded there is insufficient evidence to recommend population screening for celiac disease. This study examined the outcomes of mass screening-identified celiac disease at baseline and follow up to help inform screening practices.
Methods:
From 2019 to 2022 the mass screening study, Autoimmunity Screening for Kids, offered free testing for autoantibodies against tissue transglutaminase (TTG). TTG+ children were referred for clinical evaluation, and if diagnosed with celiac disease, via biopsy or ESPGHAN criteria, followed up after 12 months on a gluten-free diet (GFD). Demographics, clinical data, and health-related quality of life (HRQoL) at baseline and follow-up were compared. Paired parametric and non-parametric tests were applied as appropriate. For individual symptoms coded with five different levels, ordinal logistic regression with a generalized estimation equation was used to evaluate odds of improvement from baseline to follow up.
Results:
81% of enrolled children completed baseline and follow up assessments. At initial screening, 29 children reported no gastrointestinal symptoms. However, 93% reported symptoms on a more extended questionnaire, completed after knowing the screen was positive. Overall, symptoms improved on the GFD (Figure 1). At 12 months, 93% of families reported good to excellent adherence on GFD. 74% of children with low baseline ferritin improved with GFD (p<0.001). HRQoL also improved after 12 months of treatment with the GFD (Figure 2). There was no statistical difference in anthropometric, anxiety, or depression measures at follow up. Patients who did not complete follow up were more likely to be male (70%, p= 0.034), non-white race (African American and “other race,” 60%, p=0.0056), with household income <$100,000/yr (66.6%, p=0.02), insured by government-funded healthcare insurance (30% compared to 7.1% responders, p=0.033), and older (average age of 12.54 (+/- 3.84) compared to 9.45 (+/-3.99) for responders, p=0.03).
Conclusions:
Mass screening, diagnosis, and implementation of a GFD may be beneficial with respect to symptoms, iron deficiency, and quality of life. Screening and treatment did not negatively impact adherence. Future studies are needed to determine if mass screening is cost effective and how these outcomes compare to established targeted screening practices.
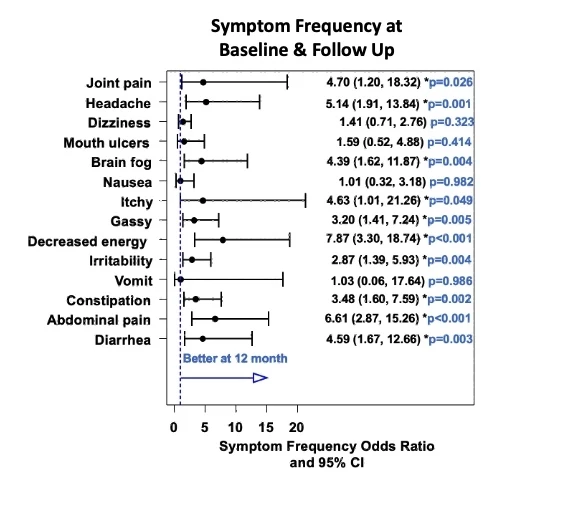
Figure 1
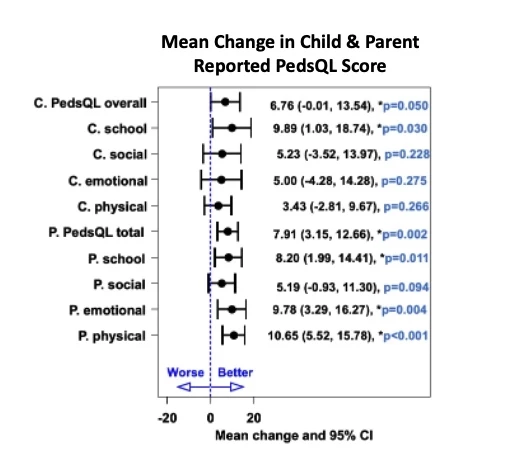
Figure 2


